See publication doi.xxxxx
ABSTRACT
Protein-truncating variants (PTVs) are a major source of genetic variation in the human genome and can profoundly affect gene function. However, their pathogenicity is often difficult to interpret due to the action of cellular surveillance mechanisms that mitigate deleterious effects. Here, we systematically analyzed more than two million PTVs from ClinVar and gnomAD across the human proteome, to assess their contribution to disease.
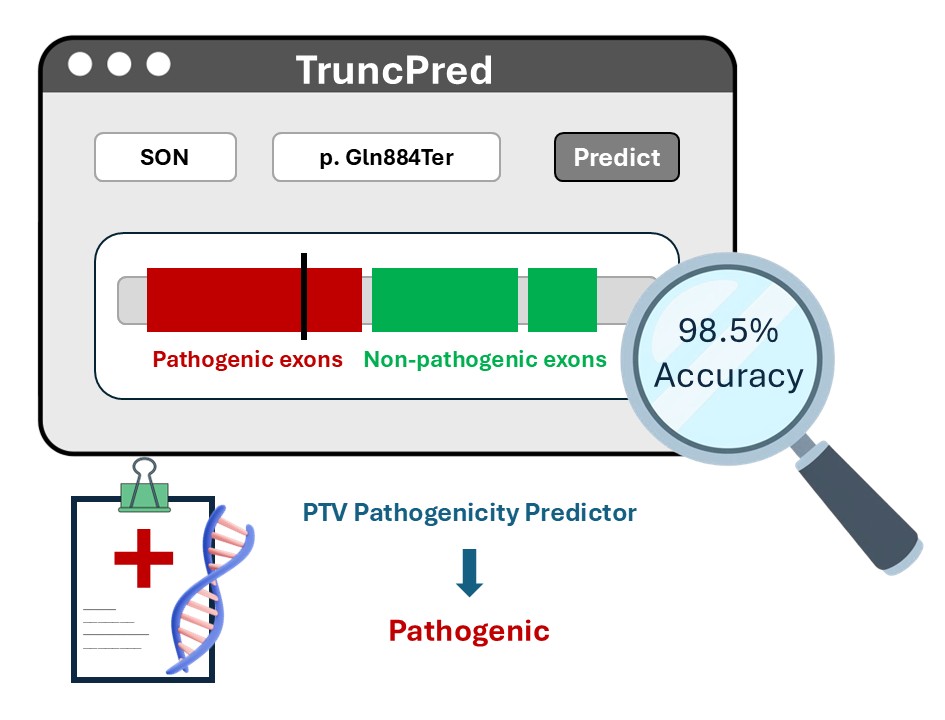
We found that the pathogenicity ratio of PTVs is substantially higher than that of missense variants. However, within autosomal-dominant (AD) proteins, only 32% of proteins that are vulnerable to missense variation also show vulnerability to PTVs, indicating that loss-of-function events are frequently buffered by cellular quality-control mechanisms such as nonsense-mediated decay (NMD). By computing exon- and domain-level pathogenicity scores, we demonstrated that 94% of all exons and 95% of all domains in AD proteins could be confidently classified as either pathogenic or non-pathogenic. When including proteins with non-Mendelian inheritance patterns, the accuracy of our model reaches 98.5%. This exon- and domain-resolved framework enables accurate prediction of the pathogenicity of novel PTVs based on their genomic location.
To translate these findings into clinical practice, we developed TruncPred (available at https://rarevariants.org/TruncPred), a web-based tool that integrates these annotations to predict the pathogenic potential of truncating variants and provide interactive visualization of their protein context.
Together, our results establish exon- and domain-level stratification as a powerful strategy for interpreting truncating variants, improving variant classification accuracy, and advancing genomic diagnostics in Mendelian disease.
This webserver and its associated code are licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. You may share and adapt the content for non-commercial purposes with proper attribution. For more details, see the full license here.
Go Back