See publication https://doi.org/10.1093/bioinformatics/btaf305
ABSTRACT
High-throughput DNA sequencing has revealed millions of single nucleotide variants (SNVs) in the human genome,
with a small fraction linked to disease. The effect of missense variants, which alter the protein sequence,
is particularly challenging to interpret due to the scarcity of clinical annotations and experimental information.
Current prediction tools, while using conservation and structural information, still struggle to predict variant pathogenicity.
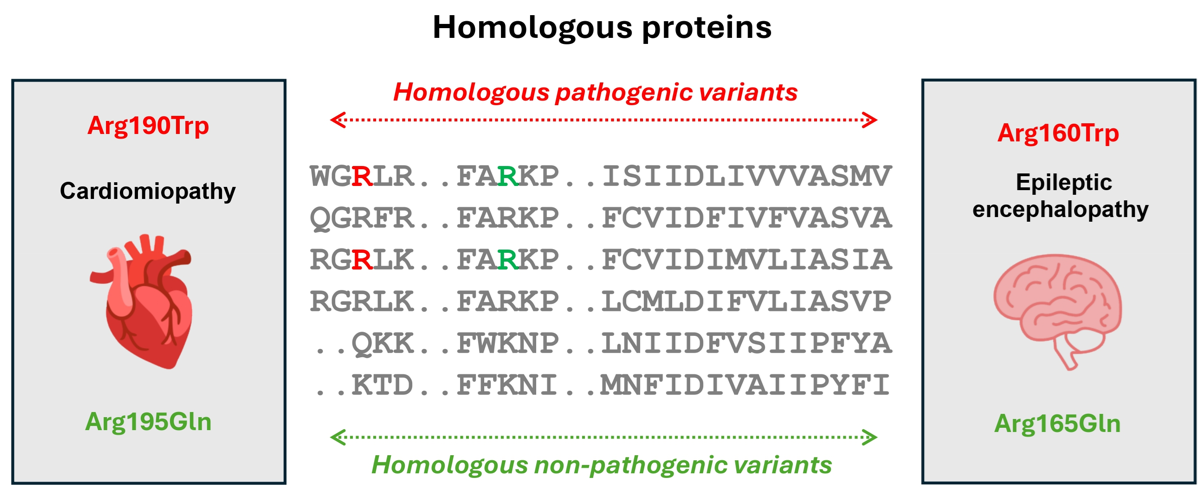
In this study, we explored the pathogenicity of homologous missense variants—variants in equivalent positions across homologous
proteins—focusing on proteins involved in autosomal dominant diseases. Our analysis of 2,976 pathogenic and 17,555 non-pathogenic
homologous variants demonstrated that pathogenicity can be extrapolated with 95% accuracy within a family, or up to 98% for closer homologs.
Moreover, we found that for the same dataset of homologous variants, the commonly used mutation predictor servers SIFT, PolyPhen-2,
and AlphaMissense were not fully capturing this biological feature. To facilitate the exploration of homologous variants, we created HomolVar,
a web server that computationally predicts the pathogenesis of missense variants using annotations from homologous variants,
freely available at https://rarevariants.org/HomolVar. Overall, these findings and the accompanying tool offer a robust method for
predicting the pathogenicity of unannotated variants, enhancing genotype-phenotype correlations, and contributing to diagnosing
rare genetic disorders.
This webserver and its associated code are licensed under the Creative Commons Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license. You may share and adapt the content for non-commercial purposes with proper attribution. For more details, see the full license here.
Go Back